rhAmp™ SNP Genotyping System
The rhAmp SNP Genotyping System is a fully integrated genotyping solution that includes an extensive predesigned assay collection, a custom design tool, reagent mixes, and optional synthetic control templates. SNP identification may be performed on any commonly available qPCR instrument.
Overview
Improve the precision of your PCR-based SNP genotyping with rhAmp SNP Genotyping technology. This technology uses a unique two-enzyme system coupled with RNA-DNA hybrid primers to interrogate target SNPs. Combined with IDT's universal reporter chemistry, rhAmp SNP Genotyping offers a simple, cost-effective genotyping solution.
- Generate quality data with greater than 99.5% call accuracy for over 90% of assays tested
- Interrogate SNPs in difficult sequence regions with amplicon lengths as short as 40 bp
- Investigate markers affordably and save on reagents using our extra-small pack size
- Gain confidence in your data with gBlocks™ Gene Fragments as control templates
The rhAmp SNP portfolio includes all components needed to generate high-quality genotyping data on the real-time PCR instrument of your choice.
rhAmp SNP Genotyping System:
- Predesigned assay collection
- Custom design tool
- Optimized reagent mixes
- Control templates
Fast and simple reaction setup
A single-tube assay setup allows for routine automation and delivers genotypes with only 90 minutes of cycling time (Figure 1). Hot-start enzymes enable benchtop reaction setup and stability of reactions for up to 2 days before cycling at room temperature.
Figure 1. Simple, one-tube reaction chemistry supports streamlined lab processes. All reagents are combined in the initial reaction setup that is stable for up to 48 hours at room temperature. Reaction setup, instrument run-time, and data acquisition may be completed in under 3 hours.
Fine-tuned chemistry for SNP genotyping
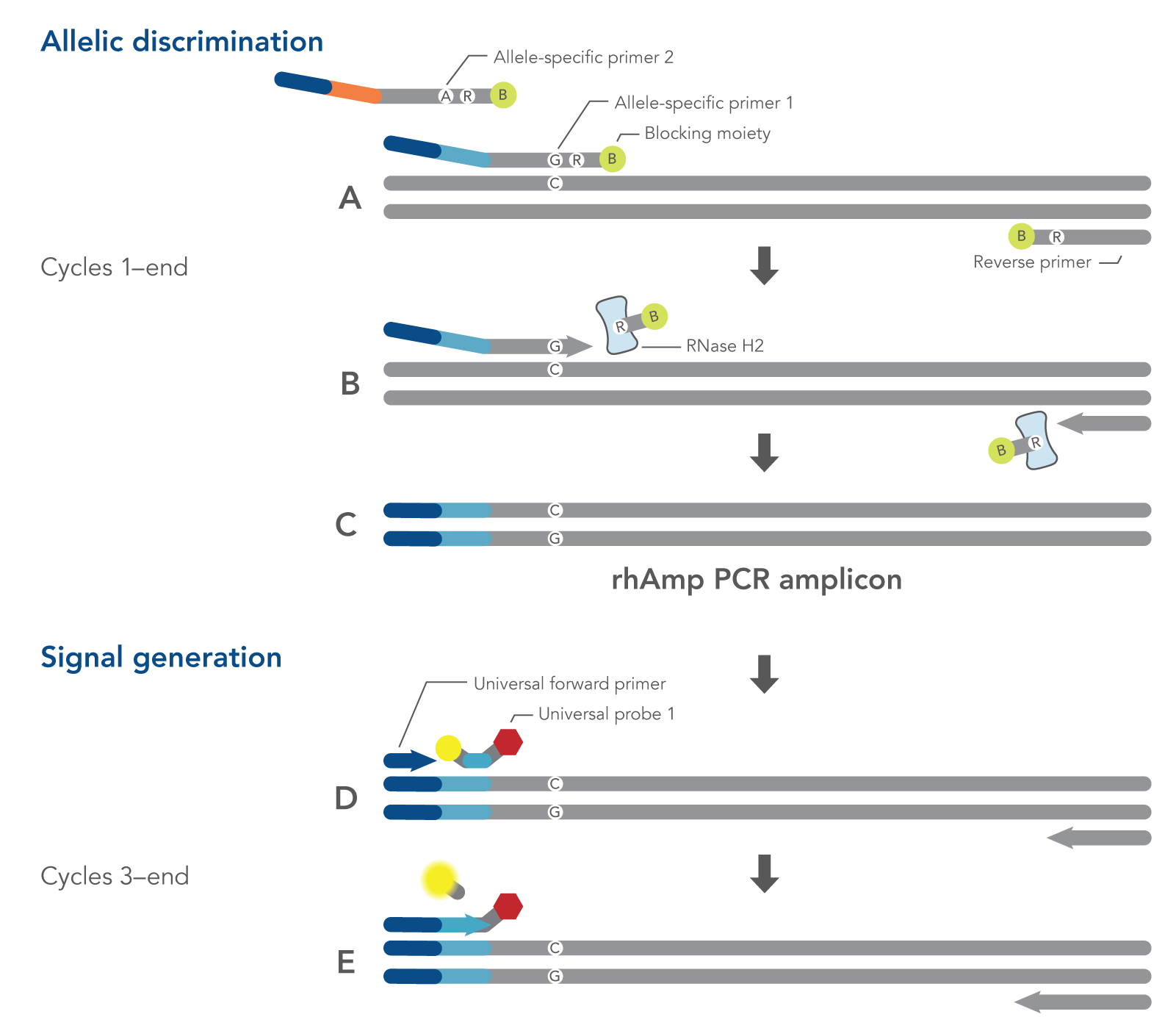
The rhAmp SNP Genotyping System offers enhanced allelic discrimination versus traditional methods. Blocked primers minimize non-specific amplification. The 3’ end of rhAmp primers incorporate a blocking group that prevents extension unless cleavage and de-blocking occur by RNase H2 enzyme (Figure 2). RNase H2 enzyme recognizes this RNA base only if it is hybridized to its perfect complement, initiating primer cleavage and activation. Learn more about rhAmp PCR technology.
Figure 2. Schematic representation of a rhAmp SNP Genotyping PCR cycle. All components needed to measure both alleles are combined in a single reaction before cycling. (A) Both allele-specific primers query the SNP locus. (B-C) RNase H2 enzyme cleaves primers that are hybridized to the perfectly matched target sequence, removing the RNA base and 3’ blocking modification, which allows extension by DNA Polymerase. (A-C) During the first two amplification cycles, a tail sequence is incorporated into the amplicon that is subsequently recognized (D) by a universal, probe-based reporter system. (E) Polymerase extension leads to degradation of the signal generation probe.